Graph theory has experienced notable growth due to its foundational role in applied mathematics and computer science, influencing fields like combinatorial optimization, biochemistry, physics, electrical engineering (particularly in communication networks and coding theory), and operational research (with scheduling applications). This paper focuses on computing topological properties, especially in molecular structures, with a specific emphasis on the nanotube \(HAC_{5}C_{7}[w,t]\).
Graph theory originated with Leonhard Euler’s solution to the Seven Bridges of Königsberg problem in 1735. It has since become a fundamental area of applied mathematics, dealing with the study of objects with connections and points known as graphs. If these connections are all one-way, the graph is termed a directed graph or digraph. Graph theory finds applications in various fields such as computer science, where it is used to model communication networks, computational devices, data organization, and computation flow. Additionally, it is employed to study molecular properties in chemistry, physics, and biology.
Claude Shannon’s seminal work laid the groundwork for exploring the information content of graphs and networks over the past half-century [1, 2]. Graph entropy has emerged as a metric to measure the structural complexity of graphs and networks [3], with applications in disciplines like information theory, biology, chemistry, and sociology.
In mathematics, graph theory entails the study of graphs, which are represented by vertices (nodes) connected by edges (lines). These mathematical structures are utilized to examine pairwise relations between objects, facilitating the construction of road maps, diagrams, and other schematics. Graphs play a crucial role in modern technological processes and communication, fostering logical and abstract thinking. For instance, connecting dots to form shapes like animals in a game represents a basic form of graph.
Chemical graph theory, a branch with around 500 articles published annually, focuses on chemical indices derived from graph theory. These indices find applications in chemistry, aiding chemists in their analyses. Prominent indices include the Zagreb indices, Wiener index, Randic index, and various connectivity indices [4, 5, 6, 7]. Topological indices, defined as graph invariants useful for chemical determinations, play a significant role in quantitative structure-property and structure-activity relationships (QSPR and QSAR) [8, 9, 10, 11, 12, 13, 14, 15].
The study of graphs extends to their relationship with finite sequences of symbols, as seen in Parikh word representable graphs (PWRGs). PWRGs are introduced based on the concept of subwords and the Parikh matrix, leading to investigations into their properties [16, 17, 18, 19, 20, 21, 22].
Furthermore, chemical graph theory has sparked interest in various topological indices associated with graphs, aiming to describe molecular structures in terms of these indices [23, 24, 25, 26, 27, 28, 29]. Numerous studies have provided formulas for computing these indices and established constraints on their values [30, 31].
This paper focuses on distance-based topological indices of binary core word PWRGs, expanding upon recent research in the field.
Topological indices are real numbers associated with compounds and graph networks, providing insights into their properties. These indices remain invariant and are crucial for predicting the properties and bioactivity of compounds in cheminformatics. They establish quantitative relationships between molecular structure and property/activity. Polynomials, such as the Wiener polynomial or Hosoya polynomial [32], are instrumental in creating distance-dependent topological indices. The M-polynomial, introduced earlier, is used for degree-dependent topological indices, reflecting the close relationship between valence and degree in chemistry [33].
Since the 1970s, degree-based graph invariants like the First and Second Zagreb indices have been widely studied. Various types of graph indices are discussed below.
Definition 1. The first \((a,b)\)-KA index, denoted \(KA^1_{(a,b)}(G)\), is defined as \[\label{e1}\tag{1} KA^1_{(a,b)}(G) = \sum_{xy \in E(G)} \left[ (d_G(x)^{a} + d_G(y)^{a}) \right]^{b},\] where \(a,b \in \mathbb{R}\) are suitably chosen parameters.
Definition 2. The reduced 1st \((a,b)\)-KA index for a graph \(G\) is defined as \[\label{e2}\tag{2} RKA^1_{a,b}(G) = \sum_{xy \in E(G)} \left[ (d_G(x)-1)^{a} + (d_G(y)-1)^{a} \right].\]
Definition 3. The reduced 2nd \((a,b)\)-KA index for a graph \(G\) is defined as \[\label{e3}\tag{3} RKA^2_{a,b}(G) = \sum_{xy \in E(G)} \left[ (d_G(x))^{a} (d_G(y))^{a} \right].\]
Definition 4. The reduced 2nd \((a,b)\)-KA index for a graph \(G\) is defined as \[ RKA^2_{a,b}(G) = \sum_{xy \in E(G)} \left[ (d_G(x)-1)^{a} (d_G(y)-1)^{a} \right]\tag{4}.\]
Definition 5. The Sombor index is introduced in [34] as \[\label{e4}\tag{5} SO(G) = \sum_{xy \in E(G)} \sqrt{(d_{G}(x)^{2} + (d_{G}(y))^{2})}.\] If we set \(a=2\) and \(b=\frac{1}{2}\) in the first \((a,b)\)-KA index, we obtain the Sombor index.
Definition 6. The modified Sombor index, denoted by \(^mSO(G)\) [35], for a graph \(G\) is defined as \[\label{e5}\tag{6} ^{m}SO(G) = \sum_{xy \in E(G)} \frac{1}{\sqrt{(d_{G}(x)^{2} + (d_{G}(y)^{2})}}.\]
Definition 7. The reduced Sombor index for a graph \(G\) can be defined as \[\label{e6}\tag{7} RSO(G) = \sum_{xy \in E(G)} \sqrt{(d_{G}(x-1)^{2} + (d_{G}(y-1)^{2})}.\]
Definition 8. The reduced modified Sombor index for a graph \(G\) can be defined as \[\label{e7}\tag{8} RSO(G) = \sum_{xy \in E(G)} \frac{1}{\sqrt{(d_{G}(x-1)^{2} + (d_{G}(y-1)^{2})}}.\]
In this section, we delve into the chemical structures of the nanotubes, specifically \(HAC_{5}C_{7}[w,t]\). By employing the edge partition of the nanotube, we evaluate several topological invariants.
Figure 1 displays the carbon nanotube \(HAC_{5}C_{7}[w;t]\)’s molecular graphs. Cycles \(C_{5}\) and \(C_{7}(C_{5}C_{7})\) net, a trivalent decoration made by alternating \(C_{5}\) and \(C_{7}\) by various chemicals, make up the architecture of this nanotube. It can cover a torus or a cylinder.
Heptagon and pentagon nets make up the structure of this nanotube. Since all \(C_{5}C_{7}\) nets are \(8\) bytes in size and there are \(wt\) heptagons \(C_{7}\) in \(H\), the size of the vertex set of the \(HAC_{5}C_{7}[w;t]\) nanotube \((w,t)\) is equal to \(n=V(HAC_{5}C_{7}[w;t])=8wt+2w\). On the other hand, the first row of \(H\) has \(w\) numbers of vertices with degree \(2\), while the last row of \(H\) contains \(w\) numbers of vertices.
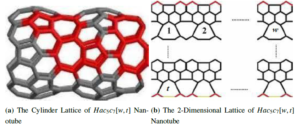
Figure 1 shows that the edge set of \(HAC_{5}C_{7}[w,t]\) may be classified into the following groups: \[\begin{aligned} E_{1}&=&\{jk\in E(HAC_{5}C_{7}[w,t]): d_{j}=2;d_{k}=2\}, \\ E_{2}&=&\{jk\in E(HAC_{5}C_{7}[w,t]):d_{j}=3;d_{k}=2\},\\ E_{3}&=&\{jk\in E(HAC_{5}C_{7}[w,t]): d_{j}=3;d_{k}=3\}. \end{aligned}\] Such that \(|E_{1}|=0;|E_{2}|=4w;|E_{3}|=12wt-2w\). We may immediately obtain the findings listed below from this edge partition.
Theorem 1. The \(KA^{1}_{(a,b)}\) of \(HAC_{5}C_{7}[w,t]\) is \[\begin{aligned} KA^{1}_{(a,b)}(HAC_{5}C_{7}[w,t])&=&\alpha w+\beta wt, \end{aligned}\] where \(\alpha= 2\left[2(3^{a}+2^{a})^{b}-(2\times 3^{a})^{b} \right]\) and \(\beta= 12\left[(2\times 3^{a} )^{b} \right]\).
Proof. Consider a chemical structure of nanotube \(HAC_{5}C_{7}[w,t]\) that is shown in Figure 1, using the edge partition of nanotube \(HAC_{5}C_{7}[w,t]\) and by using the Eq. (1), \[\begin{aligned} KA^{1}_{(a,b)}(HAC_{5}C_{7}[w,t])&=&\left|E_{1}\right|\left[2^{a}+2^{a}\right]^{b}+\left|E_{2}\right|\left[3^{a}+2^{a}\right]^{b}+\left|E_{3}\right|\left[3^{a}+3^{a}\right]^{b}\\ =0[2^{(a+1)}]^{b}+4w[3^{a}+2^{a} ]^{b}+(12wt-2w)[2\times 3^{a}]^{b}\\ =2[2(3^{a}+2^{a})^{b}-(2\times 3^{a})^{b}]w+12[(2\times 3^{a})^{b}]wt\\ =\alpha w+\beta wt, \end{aligned}\] where \(\alpha= 2[2(3^{a}+2^{a})^{b}-(2\times 3^{a})^{b} ]\) and \(\beta= 12\left[(2\times 3^{a} )^{b} \right]\). ◻
Theorem 2. The \(RKA^{1}_{(a,b)}\) of \(HAC_{5}C_{7}[w,t]\) is \[\begin{aligned} RKA^{1}_{(a,b)}(HAC_{5}C_{7}[w,t])&=& \alpha w+\beta wt, \end{aligned}\] where \(\alpha= 2\left[2(1+2^{a})^{b}-2^{(ab+b)}\right]\) and \(\beta= 3\left[2^{(ab+b+2)}\right].\)
Proof. Consider a chemical structure of nanotube \(HAC_{5}C_{7}[w,t]\) that is shown in Figure 1, using the edge partition of nanotube \(HAC_{5}C_{7}[w,t]\), by using Eq. (2), we have \[\begin{aligned} % \nonumber to remove numbering (before each equation) RKA^{1}_{(a,b)}(HAC_{5}C_{7}[w,t])&=&|E_{1}|[(2-1)^{a}+(2-1)^{a}]^{b}+|E_{2}|[(3-1)^{a}+(2-1)^{a}]^{b}\\ +|E_{3}|[(3-1)^{a}+(3-1)^{a}]^{b}\\ =0[2]^{b}+4w[1+2^{a}]^{b}+(12wt-2w)[2^{(a+1)}]^{b}\\ =2[2(1+2^{a})^{b}-2^{(ab+b)}]w+3[2^{(ab+b+2)}]wt\\ = \alpha w+\beta wt, \end{aligned}\] where \(\alpha= 2\left[2(1+2^{a})^{b}-2^{(ab+b)}\right]\) and \(\beta= 3\left[2^{(ab+b+2)}\right]\). ◻
Theorem 3. The \(KA^{2}_{(a,b)}\) of \(HAC_{5}C_{7}[w,t]\) is \[\begin{aligned} % \nonumber to remove numbering (before each equation) KA^{2}_{(a,b)}(HAC_{5}C_{7}[w,t])&=& \alpha w+\beta wt, \end{aligned}\] where \(\alpha= 2\left[2(6)^{ab}-(3)^{2ab}\right]\) and \(\beta= 4\left[(3)^{(2ab+1)}\right]\).
Proof. Consider a chemical structure of nanotube \(HAC_{5}C_{7}[w,t]\) shown in Figure 1, using the edge partition of nanotube \(HAC_{5}C_{7}[w,t]\) and by Eq. (3), we have \[\begin{aligned} % \nonumber to remove numbering (before each equation) KA^{2}_{(a,b)}(HAC_{5}C_{7}[w,t])&=&|E_{1}|[2^{a}\times 2^{a}]^{b}+|E_{2}|[3^{a}\times 2^{a}]^{b}+|E_{3}| [3^{a}\times 3^{a}]^{b}\\ &=&0[2^{2a}]^{b}+4w[6^{a}]^{b}+(12wt-2w)[3^{2a}]^{b}\\ &=&2[2(6)^{ab}-(3)^{2ab}]w+4[(3)^{(2ab+1)}]wt\\ &=&\alpha w+\beta wt, \end{aligned}\] where \(\alpha= 2\left[2(6)^{ab}-(3)^{2ab}\right]\) and \(\beta= 4\left[(3)^{(2ab+1)}\right]\). ◻
Theorem 4. The \(RKA^{2}_{(a,b)}\) index of \(HAC_{5}C_{7}[w,t]\) is \[\begin{aligned} RKA^{2}_{(a,b)}(HAC_{5}C_{7}[w,t])&=& \alpha w+\beta wt, \end{aligned}\] where, \(\alpha =2^{ab+2}-2^{2ab+1}\) and \(\beta= 3\left[2^{2(ab+1)}\right] .\)
Proof. Consider a chemical structure of nanotube \(HAC_{5}C_{7}[w,t]\) shown in Figure 1, using the edge partition of nanotube \(HAC_{5}C_{7}[w,t]\) and Eq. (4), we have \[\begin{aligned} RKA^{2}_{(a,b)}(HAC_{5}C_{7}[w,t])&=&|E_{1}|[(2-1)^{a}\times(2-1)^{a}]^{b}+|E_{2}|[(3-1)^{a}\times(2-1)^{a}]^{b}\\ &&+|E_{3}|[(3-1)^{a}\times(3-1)^{a}]^{b}\\ &=&0+4w[2^{a}]^{b}+(12wt-2w)[2^{2a}]^{b}\\ &=&\alpha w+\beta wt, \end{aligned}\] where, \(\alpha =2^{ab+2}-2^{2ab+1}\) and \(\beta= 3\left[2^{2(ab+1)}\right]\). ◻
Theorem 5. The Sombor index of \(HAC_{5}C_{7}[w,t]\) is \[\begin{aligned} S(HAC_{5}C_{7}[w,t],H_{2})&=&\alpha w+\beta wt, \end{aligned}\] where, \(\alpha =2\left[2\sqrt{13}-3\sqrt{2}\right]\) and \(\beta= 36\left[\sqrt{2}\right]\).
Proof. Consider a chemical structure of nanotube \(HAC_{5}C_{7}[w,t]\) shown in Figure 1, using the edge partition of nanotube \(HAC_{5}C_{7}[w,t]\) and Eq. (5), we have \[\begin{aligned} % \nonumber to remove numbering (before each equation) SO(HAC_{5}C_{7}[w,t])&=&|E_{1}|\sqrt{(2^{2}+2^{2})}+|E_{2}|\sqrt{(2^{2}+3^{2})}+|E_{3}|\sqrt{(3^{2}+3^{2})}\\ &=&0\sqrt{8}+4w\sqrt{13}+(12wt-2w)\sqrt{18}\\ &=&[4\sqrt{13}-6\sqrt{2}]w+36[\sqrt{2}]wt\\ &=&\alpha w+\beta wt, \end{aligned}\] where, \(\alpha =2\left[2\sqrt{13}-3\sqrt{2}\right]\) and \(\beta= 36\left[\sqrt{2}\right]\). ◻
Theorem 6. The modified Sombor index of \(HAC_{5}C_{7}[w,t]\) is \[\begin{aligned} ^{m}SO(HAC_{5}C_{7}[w,t],H_{2})&=&\alpha w+\beta wt \end{aligned}\] where, \(\alpha =\frac{4}{\sqrt{13}}-\frac{\sqrt{2}}{3}\) and \(\beta= 2\left[\sqrt{2}\right]\).
Proof. Consider a chemical structure of nanotube \(HAC_{5}C_{7}[w,t]\) shown in Figure 1, using the edge partition of nanotube \(HAC_{5}C_{7}[w,t]\) and Eq. (6), we have \[\begin{aligned} % \nonumber to remove numbering (before each equation) ^{m}SO(HAC_{5}C_{7}[w,t])&=&\frac{|E_{1}|}{\sqrt{(2^{2}+2^{2})}}+\frac{|E_{2}|}{\sqrt{(2^{2}+3^{2})}}+\frac{|E_{3}|}{\sqrt{(3^{2}+3^{2})}}\\ &=&\frac{0}{\sqrt{8}}+\frac{4w}{\sqrt{13}}+\frac{(12wt-2w)}{\sqrt{18}}\\ &=&[\frac{4}{\sqrt{13}}-\frac{\sqrt{2}}{3}]w+2[\sqrt{2}]wt\\ &=&\alpha w+\beta wt, \end{aligned}\] where, \(\alpha =\frac{4}{\sqrt{13}}-\frac{\sqrt{2}}{3}\) and \(\beta= 2\left[\sqrt{2}\right]\). ◻
Theorem 7. The reduced Sombor index of \(HAC_{5}C_{7}[w,t]\) is \[\begin{aligned} RSO(HAC_{5}C_{7}[w,t],H_{2})&=&\alpha w+\beta wt, \end{aligned}\] where, \(\alpha =4\left[\sqrt{5}-\sqrt{2}\right]\) and \(\beta= 24\left[\sqrt{2}\right]\).
Proof. Consider a chemical structure of nanotube \(HAC_{5}C_{7}[w,t]\) shown in Figure 1, using the edge partition of nanotube \(HAC_{5}C_{7}[w,t]\) and Eq. (7), we have \[\begin{aligned} % \nonumber to remove numbering (before each equation) RSO(HAC_{5}C_{7}[w,t])&=&|E_{1}|\sqrt{(2-1)^{2}+(2-1)^{2}}+|E_{2}|\sqrt{(2-1)^{2}+(3-1)^{2}}+|E_{3}|\sqrt{(3-1)^{2}+(3-1)^{2}}\\ &=&0\sqrt{2}+4w\sqrt{5}+(12wt-2w)\sqrt{8}\\ &=&4[\sqrt{5}-\sqrt{2}]w+24[\sqrt{2}]wt\\ &=&\alpha w+\beta wt, \end{aligned}\] where, \(\alpha =4\left[\sqrt{5}-\sqrt{2}\right]\) and \(\beta= 24\left[\sqrt{2}\right]\). ◻
Theorem 8. The modified reduced Sombor index of \(HAC_{5}C_{7}[w,t]\) is \[\begin{aligned} ^{m}RSO(HAC_{5}C_{7}[w,t],H_{2})&=&\alpha w+\beta wt, \end{aligned}\] where, \(\alpha =\frac{4}{\sqrt{5}}-\frac{1}{1}{\sqrt{2}}\) and \(\beta= 3\left[\sqrt{2}\right]\).
Proof. Consider a chemical structure of nanotube \(HAC_{5}C_{7}[w,t]\) shown in Figure 1, using the edge partition of nanotube \(HAC_{5}C_{7}[w,t]\) and Eq. (8), we have \[\begin{aligned} % \nonumber to remove numbering (before each equation) ^{m}RSO(HAC_{5}C_{7}[w,t])&=&\frac{|E_{1}|}{\sqrt{(2-1)^{2}+(2-1)^{2}}}+\frac{|E_{2}|}{\sqrt{(2-1)^{2}+(3-1)^{2}}}+\frac{|E_{3}|}{\sqrt{(3-1)^{2}+(3-1)^{2}}}\\ &=&\frac{0}{\sqrt{2}}+\frac{4w}{\sqrt{5}}+\frac{(12wt-2w)}{\sqrt{8}}\\ &=&[\frac{4}{\sqrt{5}}-\frac{1}{1}{\sqrt{2}}]w+3[\sqrt{2}]wt\\ &=&\alpha w+\beta wt, \end{aligned}\] where, \(\alpha =\frac{4}{\sqrt{5}}-\frac{1}{1}{\sqrt{2}}\) and \(\beta= 3\left[\sqrt{2}\right]\). ◻
In the realm of molecular studies, topological indices have emerged as indispensable tools, offering valuable numerical descriptors that encapsulate the nuances of various molecular structures. Through our in-depth exploration, we’ve calculated numerous topological properties, including, but not limited to, the 1st KA index, reduced 1st KA index, the 2nd KA index, and the reduced 2nd KA index, the Sombor index, modified Sombor index, reduced Sombor index, and modified reduced Sombor index, focusing specifically on the nanotube \(HAC_{5}C_{7}[w,t]\). A notable outcome of our research is the observation that the culmination of each theorem invariably takes a quadratic form, highlighting a consistent pattern in our findings. Moreover, the graphical representations and comparisons significantly enhanced our comprehension of these indices. Not only did these visuals serve as clarifying agents, but they also underscored the potency of the indices in delivering outstanding results. As we forge ahead in this field, the insights gained from this study reaffirm the invaluable role of topological indices in understanding molecular intricacies.
The authors declare no conflict of interest.